
Science, Vol 300, Issue 5618,
455 , 18 April 2003
[DOI: 10.1126/science.1083557]
Chromosomal Instability and Tumors Promoted by DNA
Hypomethylation Amir
Eden,1 François
Gaudet,1,3 Alpana
Waghmare,1,2 Rudolf
Jaenisch1,2*
1 Whitehead Institute for Biomedical Research,
Massachusetts Institute of Technology, Nine Cambridge Center,
Cambridge, MA 02142, USA.
2 Department of Biology,
Massachusetts Institute of Technology, Nine Cambridge Center,
Cambridge, MA 02142, USA.
3 Department of Biology,
Ludwig Maximilians University, Munich and Max Delbrück Center,
Berlin, Germany.
* To whom correspondence
should be addressed. E-mail: jaenisch@wi.mit.edu
Human tumors often display changes in DNA methylation,
including both genome-wide hypomethylation and
site-specific hypermethylation (1,
2).
In mice, DNA hypomethylation is sufficient to induce T
cell lymphomas with consistent gain of chromosome 15 (3),
indicating that genome-wide hypomethylation plays a causal
role in cancer.
To explore further the link between DNA hypomethylation and
chromosomal instability, we studied the effect of DNA
hypomethylation on tumor-prone mice carrying mutations in
both the Neurofibromatosis 1 (Nf1) and p53
tumor suppressor genes. The Nf1 and p53 genes
are closely linked on mouse chromosome 11, and double
heterozygotes carrying the mutations on the same
chromosome (NPcis) develop soft tissue sarcomas, which
show simultaneous loss of heterozygosity (LOH) of
Nf1 and p53 (4).
When we induced genomic hypomethylation in the
Nf1+/– p53+/– (NPcis) mice by
introducing a hypomorphic allele of DNA methyltransferase
1 (Dnmt1Chip/–) (3),
the mice developed sarcomas at an earlier age compared
with NPcis littermates with normal levels of DNA
methylation (Dnmt1Chip/+) (Fig.
1A). To determine whether hypomethylation promotes
the initial LOH required for tumor development in the
NPcis mice, we compared the rates of LOH in methylated and
hypomethylated primary embryonic fibroblasts. We
developed an assay to score for Nf1–/–
p53–/– cells within a population of
heterozygous cells (i.e., cells that have undergone LOH)
(fig. S1). We then used this assay in a fluctuation
analysis (5)
to calculate the rate of LOH in hypomethylated versus
methylated cells. This analysis revealed a significant
increase in LOH rate in hypomethylated cells (2.2-fold;
P = 0.01) (Fig.
1B), consistent with the hypothesis that
hypomethylation promotes tumor development in NPcis mice
by increasing the rate of LOH.
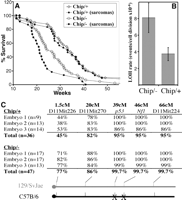 Fig. 1. (A)
Survival curve of methylated [DnmtChip/+
(Chip/+), n = 42] and hypomethylated
[Dnmt1Chip/– (Chip/–), n = 44]
Nf1+/–p53+/–cis (NPcis)
mice. Survival curve for each genotype is plotted twice: One
curve includes all mice (open symbols), and the second curve
represents only mice that developed soft tissue sarcomas
(solid symbols Chip/+), n = 33; Chip/–, n = 22).
Mice of both genotypes typically developed one predominant,
fast-growing sarcoma. Eighteen Chip/– mice developed T cell
lymphomas [as described in (3)],
including five mice that developed both a sarcoma and a
lymphoma. (B) Increased rate of LOH events in
hypomethylated mouse embryonic fibroblasts (MEFs), as
determined by fluctuation analysis (5)
(fig. S1). Graph shows the average rate in four independent
experiments (±SD). The statistical significance was determined
by Student's t test to be P = 0.01. (C)
LOH analysis. Cells in this study were derivedfrom C57/B6
x 129/svJae F1
mice. The polymorphic markers indicated were used to examine
heterozygosity along mouse chromosome 11 in cell colonies
homozygous for Nf1 and p53 derived during
fluctuation analysis. Only colonies from independent parallel
cultures were considered independent events. We analyzed
colonies obtained from three different fluctuation analyses
(n = number of independent colonies). Nf1 and
p53 mutant alleles (markedwith "x") resided on the B6
chromosome, and LOH always resulted in the loss of the 129
allele. Results are shown as the percentage of colonies with
only the B6 allele at the marked position. [View
Larger Version of this Image (44K GIF file)] Fig. 1. (A)
Survival curve of methylated [DnmtChip/+
(Chip/+), n = 42] and hypomethylated
[Dnmt1Chip/– (Chip/–), n = 44]
Nf1+/–p53+/–cis (NPcis)
mice. Survival curve for each genotype is plotted twice: One
curve includes all mice (open symbols), and the second curve
represents only mice that developed soft tissue sarcomas
(solid symbols Chip/+), n = 33; Chip/–, n = 22).
Mice of both genotypes typically developed one predominant,
fast-growing sarcoma. Eighteen Chip/– mice developed T cell
lymphomas [as described in (3)],
including five mice that developed both a sarcoma and a
lymphoma. (B) Increased rate of LOH events in
hypomethylated mouse embryonic fibroblasts (MEFs), as
determined by fluctuation analysis (5)
(fig. S1). Graph shows the average rate in four independent
experiments (±SD). The statistical significance was determined
by Student's t test to be P = 0.01. (C)
LOH analysis. Cells in this study were derivedfrom C57/B6
x 129/svJae F1
mice. The polymorphic markers indicated were used to examine
heterozygosity along mouse chromosome 11 in cell colonies
homozygous for Nf1 and p53 derived during
fluctuation analysis. Only colonies from independent parallel
cultures were considered independent events. We analyzed
colonies obtained from three different fluctuation analyses
(n = number of independent colonies). Nf1 and
p53 mutant alleles (markedwith "x") resided on the B6
chromosome, and LOH always resulted in the loss of the 129
allele. Results are shown as the percentage of colonies with
only the B6 allele at the marked position. [View
Larger Version of this Image (44K GIF file)]
|
To investigate the chromosomal mechanism leading to LOH in
the hypomethylated cells and whether specific chromosomal
regions are involved, we analyzed LOH along chromosome 11
in single colonies representing independent LOH events
(Fig.
1C). In methylated (Dnmt1Chip/+)
cells, LOH affecting the whole chromosome (including at
position 1.5 cM from the centromere) occurred in 45% of
analyzed events. The remaining 55% showed heterozygosity
at 1.5 cM but were homozygous for markers at 20 cM or 39
cM and were, therefore, the result of mitotic
recombination or of loss of the distal portion of the
chromosome. Interestingly, the frequency of LOH events
affecting the whole chromosome (including at 1.5 cM) was
significantly higher in hypomethylated cells (77% compared
with 45% in methylated cells) (Fig.
1C), suggesting that the increase in LOH rate in
hypomethylated cells is the result of a specific effect
of hypomethylation on the stability of the centromeric or
pericentric regions. A link between hypomethylation and
the stability of whole chromosome arms is also found in
the human Immunodeficiency–Centromeric Instability–Facial
Anomalies (ICF) syndrome, in hypomethylation-induced T
cell lymphomas in mice (3)
and in human hepatocellular and prostate carcinomas
(6,
7).
These results suggest that DNA hypomethylation promotes
cancer through effects on chromosomal stability. Further
characterization of the relations between DNA
methylation, chromatin composition, and chromatin
structure may allow a better understanding how DNA
hypomethylation affects chromosome structure and integrity.
References and
Notes
| 1. |
A. P. Feinberg et al., Cancer
Res. 48, 1149 (1998). |
| 2. |
P. A. Jones, S. B. Baylin, Nature
Rev. Genet. 3, 415 (2002).[ISI][Medline] |
| 3. |
F. Gaudet et al., Science
300, 455 (2003).[Free Full Text] |
| 4. |
K. Cichowski et al., Science
286, 2172
(1999).[Abstract/Free Full Text] |
| 5. |
W. S. Kendal, P. Frost, Cancer
Res. 48, 1060 (1988).[Abstract] |
| 6. |
N. Wong et al., Am. J.
Pathol. 159, 465 (2001).[Abstract/Free Full Text] |
| 7. |
W. A. Schulz et al., Genes
Chromosomes Cancer 35, 58 (2002).[CrossRef][ISI][Medline] |
| 8. |
We thank T. Jacks and K. Reilly for
NPcis mice and V. Carey for advice on biostatistics. Supported
by an EMBO fellowship ALTF 43-1999 (A.E.), by Boehringer
Ingelheim (A.E.), and by an NIH grant CA87869 (R.J.). |
Supporting Online Material
www.sciencemag.org/cgi/content/full/300/5618/455/DC1
Fig. S1
10.1126/science.1083557
Include this information when citing
this paper.
This article has been cited by other
articles:
- Geiman, T. M., Sankpal, U. T., Robertson, A. K., Chen, Y.,
Mazumdar, M., Heale, J. T., Schmiesing, J. A., Kim, W., Yokomori,
K., Zhao, Y., Robertson, K. D. (2004). Isolation and
characterization of a novel DNA methyltransferase complex linking
DNMT3B with components of the mitotic chromosome condensation
machinery. Nucleic Acids Res 32: 2716-2729 [Abstract]
[Full
Text]
- Sun, L.-Q., Lee, D. W., Zhang, Q., Xiao, W., Raabe, E. H.,
Meeker, A., Miao, D., Huso, D. L., Arceci, R. J. (2004). Growth
retardation and premature aging phenotypes in mice with disruption
of the SNF2-like gene, PASG. Genes & Dev. 18:
1035-1046 [Abstract]
[Full
Text]
- Komarova, N. L., Wodarz, D. (2004). The optimal rate of
chromosome loss for the inactivation of tumor suppressor genes in
cancer. Proc. Natl. Acad. Sci. U. S. A. 101: 7017-7021
[Abstract]
[Full
Text]
- Yanagawa, N., Tamura, G., Honda, T., Endoh, M., Nishizuka, S.,
Motoyama, T. (2004). Demethylation of the Synuclein {gamma} Gene
CpG Island in Primary Gastric Cancers and Gastric Cancer Cell
Lines. Clin Cancer Res 10: 2447-2451 [Abstract]
[Full
Text]
- Rush, L. J., Raval, A., Funchain, P., Johnson, A. J., Smith,
L., Lucas, D. M., Bembea, M., Liu, T.-H., Heerema, N. A.,
Rassenti, L., Liyanarachchi, S., Davuluri, R., Byrd, J. C., Plass,
C. (2004). Epigenetic Profiling in Chronic Lymphocytic Leukemia
Reveals Novel Methylation Targets. Cancer Res 64:
2424-2433 [Abstract]
[Full
Text]
- Wada, K., Maesawa, C., Akasaka, T., Masuda, T. (2004).
Aberrant Expression of the Maspin Gene Associated with Epigenetic
Modification in Melanoma Cells. J Invest Dermatol 122:
805-811 [Abstract]
[Full
Text]
- Liu, Z., Fisher, R. A. (2004). RGS6 Interacts with DMAP1 and
DNMT1 and Inhibits DMAP1 Transcriptional Repressor Activity.
J. Biol. Chem. 279: 14120-14128 [Abstract]
[Full
Text]
- Xin, H., Yoon, H.-G., Singh, P. B., Wong, J., Qin, J. (2004).
Components of a Pathway Maintaining Histone Modification and
Heterochromatin Protein 1 Binding at the Pericentric
Heterochromatin in Mammalian Cells. J. Biol. Chem. 279:
9539-9546 [Abstract]
[Full
Text]
- Gaudet, F., Rideout, W. M. III, Meissner, A., Dausman, J.,
Leonhardt, H., Jaenisch, R. (2004). Dnmt1 Expression in Pre- and
Postimplantation Embryogenesis and the Maintenance of IAP
Silencing. Mol. Cell. Biol. 24: 1640-1648 [Abstract]
[Full
Text]
- Karpf, A. R., Lasek, A. W., Ririe, T. O., Hanks, A. N.,
Grossman, D., Jones, D. A. (2004). Limited Gene Activation in
Tumor and Normal Epithelial Cells Treated with the DNA
Methyltransferase Inhibitor 5-Aza-2'-deoxycytidine. Mol
Pharmacol 65: 18-27 [Abstract]
[Full
Text]
- Roschke, A. V., Tonon, G., Gehlhaus, K. S., McTyre, N.,
Bussey, K. J., Lababidi, S., Scudiero, D. A., Weinstein, J. N.,
Kirsch, I. R. (2003). Karyotypic Complexity of the NCI-60
Drug-Screening Panel. Cancer Res 63: 8634-8647 [Abstract]
[Full
Text]
- Rabinowicz, P. D., Palmer, L. E., May, B. P., Hemann, M. T.,
Lowe, S. W., McCombie, W. R., Martienssen, R. A. (2003). Genes and
Transposons Are Differentially Methylated in Plants, but Not in
Mammals. Genome Res. 13: 2658-2664 [Abstract]
[Full
Text]
- Kopelovich, L., Crowell, J. A., Fay, J. R. (2003). The
Epigenome as a Target for Cancer Chemoprevention. J Natl
Cancer Inst 95: 1747-1757 [Abstract]
[Full
Text]
- Fang, M. Z., Wang, Y., Ai, N., Hou, Z., Sun, Y., Lu, H.,
Welsh, W., Yang, C. S. (2003). Tea Polyphenol
(-)-Epigallocatechin-3-Gallate Inhibits DNA Methyltransferase and
Reactivates Methylation-Silenced Genes in Cancer Cell Lines.
Cancer Res 63: 7563-7570 [Abstract]
[Full
Text]
- Herman, J. G., Baylin, S. B. (2003). Gene Silencing in Cancer
in Association with Promoter Hypermethylation. N Engl J
Med 349: 2042-2054 [Full
Text]
- Eden, A., Gaudet, F., Jaenisch, R. (2003). Response to Comment
on "Chromosomal Instability and Tumors Promoted by DNA
Hypomethylation" and "Induction of Tumors in Mice by Genomic
Hypomethylation". Science 302: 1153c-1153 [Full
Text]
- Yang, A. S., Estecio, M. R.H., Garcia-Manero, G., Kantarjian,
H. M., Issa, J.-P. J. (2003). Comment on "Chromosomal Instability
and Tumors Promoted by DNA Hypomethylation" and "Induction of
Tumors in Mice by Genomic Hypomethylation". Science 302:
1153b-1153 [Full
Text]
- Gaudet, F., Hodgson, J. G., Eden, A., Jackson-Grusby, L.,
Dausman, J., Gray, J. W., Leonhardt, H., Jaenisch, R. (2003).
Induction of Tumors in Mice by Genomic Hypomethylation.
Science 300: 489-492 [Abstract]
[Full
Text]
- Eden, A., Gaudet, F., Waghmare, A., Jaenisch, R. (2003).
Chromosomal Instability and Tumors Promoted by DNA
Hypomethylation. Science 300: 455-455 [Full
Text]
Related articles in Science:
- CANCER:
An Unstable
Liaison
- Christoph Lengauer
Science 2003 300: 442-443. (in
Perspectives) [Summary]
[Full
Text]
- Induction of Tumors in Mice by Genomic
Hypomethylation
- François Gaudet, J. Graeme Hodgson, Amir Eden, Laurie
Jackson-Grusby, Jessica Dausman, Joe W. Gray, Heinrich Leonhardt,
and Rudolf Jaenisch
Science 2003 300: 489-492. (in Reports)
[Abstract]
[Full
Text]
- Comment on "Chromosomal Instability and Tumors
Promoted by DNA Hypomethylation" and "Induction of Tumors in Mice
by Genomic Hypomethylation"
- Allen S. Yang, Marcos R.H. Estecio, Guillermo Garcia-Manero,
Hagop M. Kantarjian, and Jean-Pierre J. Issa
Science 2003 302:
1153. (in Technical Comments) [Full
Text]
- Response to Comment on "Chromosomal Instability and
Tumors Promoted by DNA Hypomethylation" and "Induction of Tumors
in Mice by Genomic Hypomethylation"
- Amir Eden, François Gaudet, and Rudolf Jaenisch
Science
2003 302: 1153. (in Technical Comments) [Full
Text]
Volume 300, Number 5618, Issue of 18 Apr 2003, p. 455.
Copyright © 2003 by The American Association for the
Advancement of Science. All rights reserved.
|

.gif)


.gif)
.gif)