 |
.gif) |
Science, Vol 302, Issue 5648, 1153 , 14 November 2003
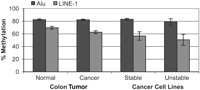
Comment on "Chromosomal Instability and Tumors Promoted by DNA Hypomethylation" and "Induction of Tumors in Mice by Genomic Hypomethylation"Eden et al. (1) and Gaudet et al. (2) reported increased tumorigenesis in mice with DNA hypomethylation and attributed these results to increased chromosomal instability. Both Gaudet et al. (2) and Lengauer (3) caution that these results could have negative implications for the use of hypomethylating agents in the treatment of cancer. However, we would like to caution against overinterpretation of these papers. It is clear that the data collected in (1, 2) are the result of rather extreme modeling in mice. The authors present data showing large decreases in repetitive element DNA methylation as a result of genetic manipulation, but these decreases may have limited relevance to human neoplasia and therapy. To address this issue, we first wished to ascertain whether the high degree of hypomethylation observed in the genetically manipulated mice is ever observed in human tumors. We examined the methylation of LINE-1 transposable elements and Alu elements using a quantitative COBRA (4) assay. PCR primers were designed based on the conserved Alu and LINE-1 sequences. Previous cloning and sequencing experiments have suggested that, in practice, the assay amplifies about 15,000 distinct Alu elements. Using this assay to quantitate global methylation, we found that in 19 pairs of colon cancer, the mean Alu methylation was identical to adjacent normal colon mucosa (normal: 82.5% methylation, SEM = 1.3%; cancer: 82.3% methylation, SEM = 1.0%; Fig. 1). The extent of Alu element methylation did vary in individual cases from normal to tumor, but 12 tumors showed an increase in Alu methylation (range = 1.2 to 7.4%), compared to only 7 tumors that showed a decrease (range = 2.1 to 12.0%). We did find hypomethylation of LINE-1 in colon cancer, but the mean decrease in methylation was only 7.0% (n = 23; range = 5.2 to 27.8%) compared to uninvolved adjacent mucosa. Normal colon mucosa had a mean LINE-1 methylation of 69.7% (SEM = 2.2%) while colon tumors showed 62.7% (SEM = 1.0%) LINE-1 methylation (Fig. 1).
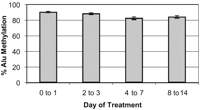
Next, we investigated the relationship between hypomethylation and chromosomal instability. Lengauer et al. (5, 6) previously reported an inverse relationship between mismatch repair (MMR) deficiency and chromosomal instability (CIN) in colon cancer cell lines. Cells that were MMR deficient used genetic instability as a method to drive tumorigenesis, and had chromosomal stability. Conversely, cells that were MMR proficient had chromosomal instability as the mechanism driving tumorigenesis. We therefore analyzed a similar cell line panel for differences in Alu element and LINE-1 methylation. Alu element methylation averaged 83.1% (SEM = 1.4%) in stable cell lines (Hct116, LoVo, SW48, RKO, DLD1) and 78.9% (SEM = 5.2%) in unstable cell lines (HT29, SW480, SW837, Colo205) (Fig. 1). LINE-1 methylation in stable versus unstable cell lines was 56.6% (SEM = 6.77%) and 50.6% (SEM = 8.76%), respectively. Thus, there was only a small, nonsignificant decrease in methylation in CIN-versus CIN+ cell lines (P = 0.48, two-tailed t-test). Gaudet et al. (2) and Lengauer (3) caution that the therapeutic use of hypomethylating agents such as 5-aza-2'deoxycytidine (5-aza-dC) could be associated with the development of secondary tumors. We have used the Alu element assay to measure demethylation induced by this drug in 41 patients with leukemia treated at different doses of 5-aza-dC. As shown in Fig. 2, 5-aza-dC resulted in an average Alu demethylation of 7.7% (SEM = 1.68, SD = 9.82%) 4 to 7 days after treatment, with 31.5% being the maximum decrease seen in a single patient. In addition, methylation returned to baseline levels within as few as 15 days of finishing treatment. Thus, 5-aza-dC-induced demethylation is at least one order of magnitude lower than that reported by Gaudet et al. (2) and Eden et al. (1). Furthermore, the pharmacologically induced demethylation appears to be very transient, with methylation returning to pretreatment levels within two weeks following therapy.
Finally, we reviewed the clinical histories of 195 patients
with leukemia treated with 5-aza-dC at the University of
Texas M.D. Anderson Cancer Center. Due to the
investigational nature of the studies, we primarily
treated patients with a poor prognosis. Fifty-three
patients survived 6 months or longer after initiation of
therapy. Of these patients, 45 have died (24 patients at
6 months to 1 year, 13 at 1 to 2 years, and 8 at
References
10.1126/science.1089523 Include this information when citing this paper.
Related articles in Science:
Volume 302, Number 5648, Issue of 14 Nov 2003, p. 1153. Copyright © 2003 by The American Association for the Advancement of Science. All rights reserved. |
| ||

 2 years
after beginning therapy). Of the 8 surviving patients, 5
are alive at 2 years, 2 at 4 years, and 1 at 5 years after
therapy began. Excluding six patients who were lost to
follow-up, all deaths were the result of the initial
disease, and no patient has developed a secondary
malignancy to date. While the follow-up is short and the
number of patients limited, we note that other drugs can
cause malignancies within months of therapy, and we
suggest that 5-aza-dC is likely to be relatively safe in
this regard. Of course, longer periods of administration
of this drug could be associated with other problems and,
indeed, some studies have found it to be mutagenic (
2 years
after beginning therapy). Of the 8 surviving patients, 5
are alive at 2 years, 2 at 4 years, and 1 at 5 years after
therapy began. Excluding six patients who were lost to
follow-up, all deaths were the result of the initial
disease, and no patient has developed a secondary
malignancy to date. While the follow-up is short and the
number of patients limited, we note that other drugs can
cause malignancies within months of therapy, and we
suggest that 5-aza-dC is likely to be relatively safe in
this regard. Of course, longer periods of administration
of this drug could be associated with other problems and,
indeed, some studies have found it to be mutagenic (.gif)
.gif)