|
20.1 Introduction |
|
Key terms |
|
Enhancer element is a cis-acting sequence that increases the utilization of (some) eukaryotic promoters, and can function in either orientation and in any location (upstream or downstream) relative to the promoter.Response element is a sequence in a eukaryotic promoter that is recognized by a specific transcription factor. |
Initiation of transcription requires the enzyme RNA polymerase and transcription factors. Any protein that is needed for the initiation of transcription, but which is not itself part of RNA polymerase, is defined as a transcription factor. Many transcription factors act by recognizing cis-acting sites on DNA. However, binding to DNA is not the only means of action for a transcription factor. A factor may recognize another factor, or may recognize RNA polymerase, or may be incorporated into an initiation complex only in the presence of several other proteins. The ultimate test for membership of the transcription apparatus is functional: a protein must be needed for transcription to occur at a specific promoter or set of promoters.
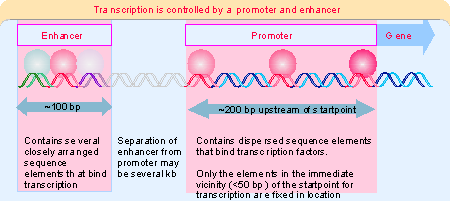
A significant difference in the transcription of eukaryotic and prokaryotic mRNAs is that initiation at a eukaryotic promoter involves a large number of factors that bind to a variety of cis-acting elements. The promoter is defined as the region containing all these binding sites, that is, which can support transcription at the normal efficiency and with the proper control. So the major feature defining the promoter for a eukaryotic mRNA is the location of binding sites for transcription factors. RNA polymerase itself binds around the startpoint, but does not directly contact the extended upstream region of the promoter. By contrast, the bacterial promoters discussed in
9 Transcription are largely defined in terms of the binding site for RNA polymerase in the immediate vicinity of the startpoint.Transcription in eukaryotic cells is divided into three classes. Each class is transcribed by a different RNA polymerase:
Accessory factors are needed for initiation, but are not required subsequently. In each case, the factors, rather than the enzymes themselves, are principally responsible for recognizing the promoter. This is different from bacterial RNA polymerase, where it is the enzyme that recognizes the promoter sequences.
The promoters for RNA polymerases I and II are (mostly) upstream of the startpoint, but some promoters for RNA polymerase III lie downstream of the startpoint. Each promoter contains characteristic sets of short conserved sequences that are recognized by the appropriate class of factors. RNA polymerases I and III each recognize a relatively restricted set of promoters, and rely upon a small number of accessory factors.
Promoters utilized by RNA polymerase II show more variation in sequence, and have a modular. Short sequence elements that are recognized by transcription factors lie upstream of the startpoint. These cis-acting sites usually are spread out over a region of >200 bp. Some of these elements and the factors that recognize them are common: they are found in a variety of promoters and are used constitutively. Others are specific: they identify particular classes of genes and their use is regulated. The elements occur in different combinations in individual promoters.
Many factors are used by RNA polymerase II. They fall into three groups with regard to mechanism of action:
All RNA polymerase II promoters have similar sequences that are bound by the basal apparatus and that establish the site of initiation. The sequences upstream determine whether the promoter is expressed in all cell types or is specifically regulated. Promoters that are constitutively expressed (their genes are sometimes called housekeeping genes) have upstream sequence elements that are recognized by ubiquitous activators. No element/factor combination is an essential component of the promoter, which suggests that initiation by RNA polymerase II may be sponsored in many different ways. Promoters that are expressed only in certain times or places have response elements that require activators that are available only at those times or places.
Sequence components of the promoter are defined operationally by the demand that they must be located in the general vicinity of the startpoint and are required for initiation. The
enhancer is another type of site involved in initiation. It is identified by sequences that stimulate initiation, but that are located a considerable distance from the startpoint. Enhancer elements are often targets for tissue-specific or temporal regulation. Figure 20.1 illustrates the general properties of promoters and enhancers.The components of an enhancer resemble those of the promoter; they consist of a variety of modular elements. However, the elements are organized in a closely packed array. The elements in an enhancer function like those in the promoter, but the enhancer does not need to be near the startpoint. However, proteins bound at enhancer elements interact with proteins bound at promoter elements. The distinction between promoters and enhancers is operational, rather than implying a fundamental difference in mechanism. This view is fortified by the fact that some types of element are found in both promoters and enhancers.
The common mode of regulation of eukaryotic transcription is positive: a transcription factor is provided under tissue-specific control to activate a promoter or set of promoters that contain a common target sequence. Regulation by specific repression of a target promoter is less common.
A eukaryotic transcription unit generally contains a single gene, and termination occurs beyond the end of the coding region. We should like to define the mechanism of termination, but it lacks the regulatory importance that applies in prokaryotic systems. RNA polymerases I and III terminate at discrete sequences in defined reactions, but the mode of termination by RNA polymerase II is not clear. However, the significant event in generating the 3![]() end of an mRNA is not the termination event itself, but instead results from a cleavage reaction in the primary transcript (see
end of an mRNA is not the termination event itself, but instead results from a cleavage reaction in the primary transcript (see
This section updated 4-30-2001
|
20.2 Eukaryotic RNA polymerases consist of many subunits |
|
Key terms |
|
Amanitin (more fully a-amanitin) is a bicyclic octapeptide derived from the poisonous mushroom Amanita phalloides; it inhibits transcription by certain eukaryotic RNA polymerases, especially RNA polymerase II. |
|
Key Concepts |
|
· RNA polymerase I synthesizes rRNA in the nucleolus.· RNA polymerase II synthesizes mRNA in the nucleoplasm.· RNA polymerase III synthesizes small RNAs in the nucleoplasm.· All eukaryotic RNA polymerases have ~12 subunits and are aggregates of >500 kD.· Some subunits are common to all three RNA polymerases.· The largest subunit in RNA polymerase II has a CTD (carboxy-terminal domain) consisting of multiple repeats of a septamer. |
The three eukaryotic RNA polymerases have different locations in the nucleus, corresponding with the genes that they transcribe (for review see
71).The most prominent activity is the enzyme RNA polymerase I, which resides in the nucleolus and is responsible for transcribing the genes coding for rRNA. It accounts for most cellular RNA synthesis.
The other major enzyme is RNA polymerase II, located in the nucleoplasm (the part of the nucleus excluding the nucleolus). It represents most of the remaining cellular activity and is responsible for synthesizing heterogeneous nuclear RNA (hnRNA), the precursor for mRNA.
RNA polymerase III is a minor enzyme activity. This nucleoplasmic enzyme synthesizes tRNAs and other small RNAs.
All eukaryotic RNA polymerases are large proteins, appearing as aggregates of >500 kD. They typically have ~12 subunits. The purified enzyme can undertake template-dependent transcription of RNA, but is not able to initiate selectively at promoters. The general constitution of a eukaryotic RNA polymerase II enzyme as typified in S. cerevisiae is illustrated in
Figure 20.2. The two largest subunits are homologous to theTen subunits of the yeast RNA polymerase II have been located on the crystal structure, as shown in
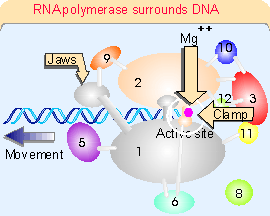
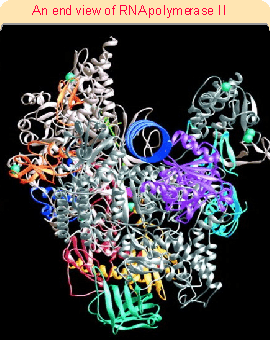
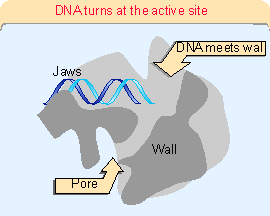
Figure 20.3 (1714). The catalytic site is formed by a cleft between the two large subunits (#1 and #2), which hold DNA downstream in "jaws". The structure is generally similar to that of bacterial RNA polymerase. This can be seen more clearly in the crystal structure of Figure 20.4. RNA polymerase surrounds the DNA, as seen in the view of Figure 20.5. A catalytic Mg2+ ion is found at the active site. The DNA is clamped in position at the active site by subunits 1, 2, and 6. Access of nucleotides to the active site from above (in the orientation of Figure 20.5) is probably blocked by the DNA, and they may come from below via pores through the structure. Figure 20.6 shows that DNA is forced to take a turn at the entrance to the site, because of adjacent wall of protein. Subunits 4 and 7 are missing from this structure; they form a subcomplex that dissociates from the complete enzyme.The largest subunit in RNA polymerase II has a carboxy-terminal domain (
CTD), which consists of multiple repeats of a consensus sequence of 7 amino acids. The sequence is unique to RNA polymerase II. There are ~26 repeats in yeast and ~50 in mammals. The number of repeats is important, because deletions that remove (typically) more than half of the repeats are lethal (in yeast). The CTD can be highly phosphorylated on serine or threonine residues; this is involved in the initiation reaction (see later).The RNA polymerase activities of mitochondria and chloroplasts are smaller, and resemble bacterial RNA polymerase rather than any of the nuclear enzymes. Of course, the organelle genomes are much smaller, the resident polymerase needs to transcribe relatively few genes, and the control of transcription is likely to be very much simpler (if existing at all). So these enzymes are analogous to the phage enzymes that have a single fixed purpose and do not need the ability to respond to a more complex environment.
A major practical distinction between the eukaryotic enzymes is drawn from their response to the bicyclic octapeptide ![]() -
-
This section updated 4-30-2001
|
Reviews |
|
|
71: |
Doi, R. H. and Wang, L.-F. (1986). Multiple prokaryotic RNA polymerase sigma factors. Microbiol. Rev. 50, 227-243. |
|
222: |
Young, R. A. (1991). RNA polymerase II. Ann. Rev. Biochem. 60, 689-715. |
|
Research |
|
|
1714: |
Cramer, P. , Bushnell, D. A. , Fu, J. , Gnatt, A. L. , Maier-Davis, B. , Thompson, N. E. , Burgess, R. R. , Edwards, A. M. , David, P. R. , and Kornberg, R. D. (2000). Architecture of RNA polymerase II and implications for the transcription mechanism. Science 288, 640-649. |


|
20.3 Promoter elements are defined by mutations and footprinting |
|
Key Concepts |
|
· Promoters are defined by their ability to cause transcription of an attached sequence in an appropriate test system in vitro or in vivo. |
The first step in characterizing a promoter is to define the overall length of DNA that contains all the necessary sequence elements. To do this, we need a test system in which the promoter is responsible for the production of an easily assayed product. Historically, several types of systems have been used:
When a promoter is analyzed, it is imporant that only the promoter sequence changes.
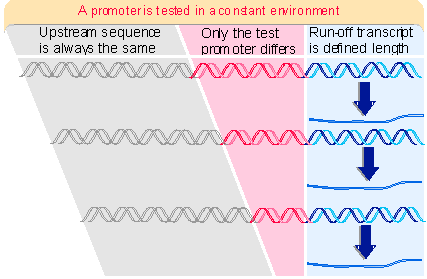
Figure 20.7 shows that the same long upstream sequence is always placed next to the promoter to ensure that it is always in the same context, . Because termination does not occur properly in the in vitro systems, the template is cut at some distance from the promoter (usually ~500 bp downstream), to ensure that all polymerases "run off" at the same point, generating an identifiable transcript.We start with a particular fragment of DNA that can initiate transcription in one of these systems. Then the boundaries of the sequence constituting the promoter can be determined by reducing the length of the fragment from either end, until at some point it ceases to be active, as illustrated in
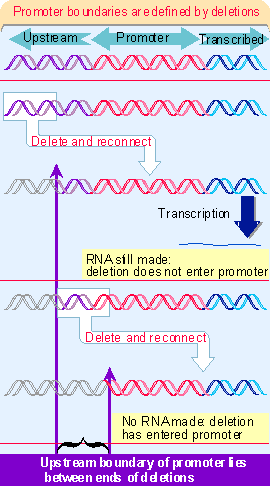
Figure 20.8. The boundary upstream can be identified by progressively removing material from this end until promoter function is lost. To test the boundary downstream, it is necessary to reconnect the shortened promoter to the sequence to be transcribed (since otherwise there is no product to assay).Once the boundaries of the promoter have been defined, the importance of particular bases within it can be determined by introducing point mutations or other rearrangements in the sequence. As with bacterial RNA polymerase, these can be characterized as up or down mutations. Some of these rearrangements affect only the rate of initiation; others influence the site at which initiation occurs, as seen in a change of the startpoint. To be sure that we are dealing with comparable products, in each case it is necessary to characterize the 5![]() end of the RNA.
end of the RNA.
We can apply several criteria in identifying the sequence components of a promoter (or any other site in DNA):
This section updated 4-30-2001
|
20.4 RNA polymerase I has a bipartite promoter |
|
Key Concepts |
|
· The RNA polymerase I promoter consists of a core promoter and an upstream control element.· The factor UBF1 binds to both regions and enables the factor SL1 to bind.· SL1 includes the factor TBP that is involved in initiation by all three RNA polymerases.· RNA polymerase binds to the UBF1-SL1 complex at the core promoter. |
RNA polymerase I transcribes only the genes for ribosomal RNA, from a single type of promoter. The transcript includes the sequences of both large and small rRNAs, which are later released by cleavages and processing. There are many copies of the transcription unit, alternating with nontranscribed
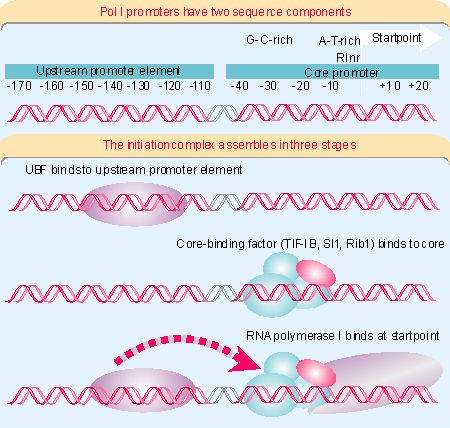
spacers, and organized in a cluster as discussed in 4 Clusters and repeats. The organization of the promoter, and the events involved in initiation, are illustrated in Figure 20.9.The promoter consists of two separate regions. The
core promoter surrounds the startpoint, extending from –45 to +20, and is sufficient for transcription to initiate. It is generally G·C-rich (unusual for a promoter) except for the only conseved sequence element, a short A·T-rich sequence around the startpoint called the RInr. However, its efficiency is very much increased by the upstream promoter element (UPE), another G·C-rich sequence, related to the core promoter sequence, and which extends from –180 to –107. This type of organization is common to pol I promoters in many species, although the actual sequences vary widely (for review see 1672).RNA polymerase I requires two ancillary factors. The factor that binds to the core promoter consists of 4 proteins. (It is called SL1, TIF-IB, Rib1 in different species). One of its components, called TBP, is a factor that is required also for initiation by RNA polymerases II and III. We discuss its role in initiation by those polymerases shortly. TBP does not bind directly to G·C-rich DNA, so DNA-binding is probably the responsibility of the other components of the core-binding factor. It is likely that TBP interacts with RNA polymerase, possibly with a common subunit or a feature that has been conserved among polymerases. Core-binding factor enables RNA polymerase I to initiate from the promoter at a low basal frequency.
The core-binding factor has primary responsibility for ensuring that the RNA polymerase is properly localized at the startpoint. We see shortly that a comparable function is provided for RNA polymerases II and III by a factor that consists of TBP associated with other proteins. So a common feature in initiation by all three polymerases is a reliance on a "positioning" factor that consists of TBP associated with proteins that are specific for each type of promoter.
For high frequency initiation, the factor UBF is required. This is a single polypeptide that binds to a G·C-rich element in the upstream promoter element. One indication of how UBF interacts with the core-binding factor is given by the importance of the spacing between the upstream promoter element and the core promoter. This can be changed by distances involving integral numbers of turns of DNA, but not by distances that introduce half turns. This implies that UBF and core-binding factor need to be bound on the same face of DNA in order to interact. In the presence of UBF, core-binding factor binds more efficiently to the core promoter (
1676).This section updated 4-30-2001
|
Reviews |
|
|
1672: |
Paule, M. R. and White, R. J. (2000). Survey and summary: transcription by RNA polymerases I and III. Nucleic Acids Res. 28, 1283-1298. |
|
Research |
|
|
1676: |
Bell, S. P. , Learned, R. M. , Jantzen, H. M. , and Tjian, R. (1988). Functional cooperativity between transcription factors UBF1 and SL1 mediates human ribosomal RNA synthesis. Science 241, 1192-1197. |
|
20.5 RNA polymerase III uses both downstream and upstream promoters |
|
Key terms |
|
Preinitiation complex in eukaryotic transcription describes the assembly of transcription factors at the promoter before RNA polymerase binds. |
|
Key Concepts |
|
· RNA polymerase III has two types of promoters.· Internal promoters have short consensus sequences located within the transcription unit and cause initiation to occur a fixed distance upstream.· TFIIIA and TFIIIC bind to the consensus sequences and enable TFIIB to bind at the startpoint.· Upstream promoters contain three short consensus sequences upstream of the startpoint that are bound by transcription factors.· For both types of promoter, the factor bound at the startpoint has TBP as one subunit and enables RNA polymerase to bind. |
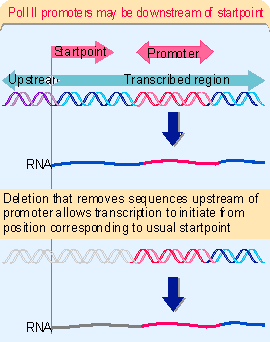
Recognition of promoters by RNA polymerase III strikingly illustrates the relative roles of transcription factors and the polymerase enzyme. The promoters fall into two general classes that are recognized in different ways by different groups of factors. The promoters for 5S and tRNA genes are internal; they lie downstream of the startpoint. The promoters for snRNA (small nuclear RNA) genes lie upstream of the startpoint in the more conventional manner of other promoters. In both cases, the individual elements that are necessary for promoter function consist exclusively of sequences recognized by transcription factors, which in turn direct the binding of RNA polymerase.
Before the promoter of 5S RNA genes was identified in X. laevis, all attempts to identify promoter sequences assumed that they would lie upstream of the startpoint. But deletion analysis showed that the 5S RNA product continues to be synthesized when the entire sequence upstream of the gene is removed!
When the deletions continue into the gene, a product very similar in size to the usual 5S RNA continues to be synthesized so long as the deletion ends before base +55.
Figure 20.10 shows that the first part of the RNA product corresponds to plasmid DNA; the second part represents the segment remaining of the usual 5S RNA sequence. But when the deletion extends past +55, transcription does not occur. So the promoter lies downstream of position +55, but causes RNA polymerase III to initiate transcription a more or less fixed distance away.When deletions extend into the gene from its distal end, transcription is unaffected so long as the first 80 bp remain intact. Once the deletion cuts into this region, transcription ceases. This places the downstream boundary position of the promoter at about position +80.
So the promoter for 5S RNA transcription lies between positions +55 and +80 within the gene. A fragment containing this region can sponsor initiation of any DNA in which it is placed, from a startpoint ~55 bp farther upstream. (The wild-type startpoint is unique; in deletions that lack it, transcription initiates at the purine base nearest to the position 55 bp upstream of the promoter. (
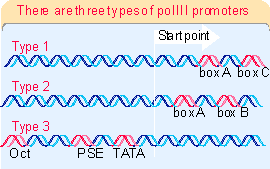
641, 642))The structures of three types of promoters for RNA polymerase III are summarized in
Figure 20.11. There are two types of internal promoter. Each contains a bipartite structure, in which two short sequence elements are separated by a variable sequence. Type 1 consists of a boxA sequence separated from a boxC sequence (1673), and type 2 consists of a boxA sequence separated from a boxB sequence (1674). The distance between boxA and boxB in a type 2 promoter can vary quite extensively, but the boxes usually cannot be brought too close together without abolishing function. Type 3 promoters have 3 sequences elements all located upstream of the startpoint (1675).The detailed interactions are different at the two types of internal promoter, but the principle is the same. TFIIIC binds downstream of the startpoint, either independently (type 2 promoters) or in conjunction with TFIIIA (type 1 promoters). The presence of TFIIIC enables the positioning factor TFIIIB to bind at the startpoint. Then RNA polymerase is recruited.
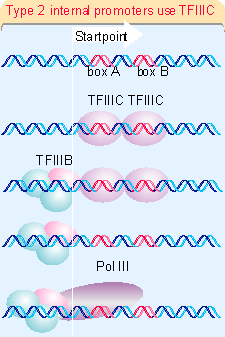
Figure 20.12 summarizes the stages of reaction at type 2 internal promoters. TFIIIC binds to both boxA and boxB. This enables TFIIIB to bind at the startpoint. Then RNA polymerase III can bind.
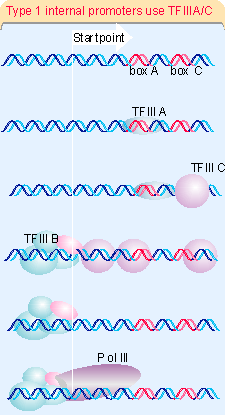
The difference at type 1 internal promoters is that TFIIIA must bind at boxA to enable TFIIIC to bind at boxC. Figure 20.13 shows that, once TFIIIC has bound, events follow the same course as at type 2 promoters, with TFIIIB binding at the startpoint, and RNA polymerase III joining the complex. Type 1 promoters are found only in the genes for 5S rRNA.
TFIIIA and TFIIIC are assembly factors, whose sole role is to assist the binding of TFIIIB at the right location. Once TFIIIB has bound, TFIIIA and TFIIIC can be removed from the promoter (by high salt concentration in vitro) without affecting the initiation reaction. TFIIIB remains bound in the vicinity of the startpoint and its presence is sufficient to allow RNA polymerase III to bind at the startpoint. So TFIIIB is the only true initiation factor required by RNA polymerase III (643). This sequence of events explains how the promoter boxes downstream can cause RNA polymerase to bind at the startpoint, farther upstream. Although the ability to transcribe these genes is conferred by the internal promoter, changes in the region immediately upstream of the startpoint can alter the efficiency of transcription.
TFIIIC is a large protein complex (>500 kD), comparable in size to RNA polymerase itself, and containing 6 subunits. TFIIIA is a member of an interesting class of zinc finger proteins that we discuss later. The positioning factor, TFIIIIB, consists of three subunits. It includes the same protein, TBP, that is present in the core-binding factor for pol I promoters, and also in the corresponding transcription factor (TFIID) for RNA polymerase II (1677). It also contains Brf, which is related to the factor TFIIB that is used by RNA polymerase II. The third subunit is called B"; it is dispensable if the DNA duplex is partially melted, which suggests that its function is to initiate the transcription bubble (945). The role of B" may be comparable to the role played by sigma factor in bacterial RNA polymerase (see 9.15 Substitution of sigma factors may control initiation).
The upstream region has a conventional role in the third class of polymerase III promoters. In the example shown in
Figure 20.11, there are three upstream elements. These elements are also found in promoters for snRNA genes that are transcribed by RNA polymerase II. (Genes for some snRNAs are transcribed by RNA polymerase II, while others are transcribed by RNA polymerase III.) The upstream elements function in a similar manner in promoters for both polymerases II and III.Initiation at an upstream promoter for RNA polymerase III can occur on a short region that immediately precedes the startpoint and contains only the TATA element. However, efficiency of transcription is much increased by the presence of the PSE and OCT elements. The factors that bind at these elements interact cooperatively. (The PSE element may be essential at promoters used by RNA polymerase II, whereas it is stimulatory in promoters used by RNA polymerase III; its name stands for proximal sequence element.)
The TATA element appears to confer specificity for the type of polymerase (II or III) that is recognized by an snRNA promoter. It is recognized by a factor that includes the TBP, which actually recognizes the sequence in DNA. The TBP is associated with other proteins, which are specific for the type of promoter. The function of TBP and its associated proteins is to position the RNA polymerase correctly at the startpoint. We discuss this in more detail for RNA polymerase II in the next section.
The factors work in the same way for both types of promoters for RNA polymerase III. The factors bind at the promoter before RNA polymerase itself can bind. They form a
preinitiation complex that directs binding of the RNA polymerase. RNA polymerase III does not itself recognizes the promoter sequence, but binds adjacent to factors that are themselves bound just upstream of the startpoint. For the type 1 and type 2 internal promoters, the assembly factors ensure that TFIIIB (which includes TBP) is bound just upstream of the startpoint, to provide the positioning information. For the upstream promoters, transcription factors that directly recognize the upstream sites form a complex (including TBP) that is recognized by RNA polymerase III. So irrespective of the location of the promoter sequences, factor(s) are bound close to the startpoint in order to direct binding of RNA polymerase III (for review see 220).This section updated 4-30-2001
|
Reviews |
|
|
220: |
Geiduschek, E. P. and Tocchini-Valentini, G. P. (1988). Transcription by RNA polymerase III. Ann. Rev. Biochem. 57, 873-914. |
|
Research |
|
|
641: |
Sakonju, S., Bogenhagen, D. F., and Brown, D. D. (1980). A control region in the center of the 5S RNA gene directs specific initiation of transcription: I the 5' border of the region. Cell 19, 13-25. |
|
642: |
Bogenhagen, D. F., Sakonju, S., and Brown, D. D. (1980). A control region in the center of the 5S RNA gene directs specific initiation of transcription: II the 3' border of the region. Cell 19, 27-35. |
|
643: |
Kassavatis, G. A., Braun, B. R., Nguyen, L. H., and Geiduschek, E. P. (1990). S. cerevisiae TFIIIB is the transcription initiation factor proper of RNA polymerase III, while TFIIIA and TFIIIC are assembly factors. Cell 60, 235-245. |
|
945: |
Kassavetis, G. A., Letts, G. A., and Geiduschek, E. P. (1999). A minimal RNA polymerase III transcription system. EMBO J. 18, 5042-5051. |
|
1673: |
Pieler, T. , Hamm, J. , and Roeder, R. G. (1987). The 5S gene internal control region is composed of three distinct sequence elements, organized as two functional domains with variable spacing. Cell 48, 91-100. |
|
1674: |
Galli, G. , Hofstetter, H. , and Birnstiel, M. L. (1981). Two conserved sequence blocks within eukaryotic tRNA genes are major promoter elements. Nature 294, 626-631. |
|
1675: |
Kunkel, G. R. and Pederson, T. (1988). Upstream elements required for efficient transcription of a human U6 RNA gene resemble those of U1 and U2 genes even though a different polymerase is used. Genes Dev. 2, 196-204. |
|
1677: |
Kassavetis, G. A. , Joazeiro, C. A. , Pisano, M. , Geiduschek, E. P. , Colbert, T. , Hahn, S. , and Blanco, J. A. (1992). The role of the TATA-binding protein in the assembly and function of the multisubunit yeast RNA polymerase III transcription factor, TFIIIB. Cell 71, 1055-1064. |